Fetching data from a CATMAID server¶
With minor exceptions, pymaid covers every API endpoint of your CATMAID server
- i.e. you should be able to get all the data that your web browser shows (and more)
pymaid has >50 different functions that fetch various kinds of data from
CATMAID. Too many to cover them all - instead this tutorial will try to give
you a flavour by showcasing a few examples.
If you know what data you are after, head over to the API reference and search for keywords to find the appropriate function.
A quick primer¶
All functions that require neurons/skeleton IDs as inputs (e.g. get_neuron()) accept either:
Skeleton ID(s) (int or str):
n = pymaid.get_neuron(16) nl = pymaid.get_neuron([16, 57241])
Neuron name(s) (str):
n = pymaid.get_neuron('PN glomerulus DC3 57242 ML') nl = pymaid.get_neuron(['PN glomerulus DC3 57242 ML', 'PN glomerulus VA6 017 DB'])Annotation(s) (str):
n = pymaid.get_neuron('annotation:glomerulus VA6') nl = pymaid.get_neuron(['annotation:glomerulus DC3', 'annotation:glomerulus VA6'])CatmaidNeuronorCatmaidNeuronListobject:n = pymaid.get_neuron(16) cn_table = pymaid.get_partners(n)
By default, annotations and neuron names must match exactly. You can use regex on names and annotations, using a leading /:
# Get all neurons that have 'VA6' in their name
nl = pymaid.get_neuron('/VA6')
# Get all neurons annotated with a 'VA6'-containing annotation
nl = pymaid.get_neuron('annotation:/VA6')
# Get all skeleton IDs that have an annotation starting with 'AV1.R':
skids = pymaid.get_skids_by_annotation('/^AV1.R')
See here for detailed explanation of regex patterns.
Fetching neurons¶
One of the most basic thing is fetching neurons. First, we need to set up the connection to the CATMAID server. This should already look familiar - if not check out the quickstart tutorial <tutorial> first:
[2]:
import pymaid
rm = pymaid.connect_catmaid()
INFO : Global CATMAID instance set. Caching is ON. (pymaid)
Now that’s done, we can get going. In 99% of the cases, you will use pymaid.get_neurons() to fetch skeletons for analysis and visualization.
[14]:
da1 = pymaid.get_neuron('annotation:glomerulus DA1 right')
da1
[14]:
| type | name | skeleton_id | n_nodes | n_connectors | n_branches | n_leafs | cable_length | soma | units | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CatmaidNeuron | Uniglomerular mALT DA1 lPN 57316 2863105 ML | 2863104 | 6774 | 470 | 280 | None | 1522064.50 | [3245741] | 1 nanometer |
| 1 | CatmaidNeuron | Uniglomerular mALT DA1 lPN 57382 ML | 57381 | 8243 | 382 | 195 | None | 1337348.75 | [3364368] | 1 nanometer |
| 2 | CatmaidNeuron | Uniglomerular mALT DA1 lPN 61222 AJ | 61221 | 8578 | 556 | 199 | None | 1353478.75 | [3254492] | 1 nanometer |
| 3 | CatmaidNeuron | Uniglomerular mALT DA1 lPN 57354 GA | 57353 | 6985 | 370 | 225 | None | 1539364.25 | [3254736] | 1 nanometer |
| 4 | CatmaidNeuron | Uniglomerular mALT DA1 lPN 57324 LK JSL | 57323 | 6223 | 481 | 256 | None | 1483462.50 | [3251355] | 1 nanometer |
| 5 | CatmaidNeuron | Uniglomerular mALT DA1 lPN 57358 IJA | 755022 | 7453 | 515 | 230 | None | 1401414.75 | [3252148] | 1 nanometer |
| 6 | CatmaidNeuron | Uniglomerular mALT DA1 lPN 27296 BH | 27295 | 10216 | 507 | 304 | None | 1806000.25 | [3005291] | 1 nanometer |
| 7 | CatmaidNeuron | Uniglomerular mlALT DA1 vPN mlALTed Milk 18114... | 1811442 | 11769 | 804 | 378 | None | 2169807.25 | [6838672] | 1 nanometer |
| 8 | CatmaidNeuron | Uniglomerular mALT DA1 lPN 57312 LK | 57311 | 5543 | 463 | 211 | None | 1364103.25 | [3059181] | 1 nanometer |
[26]:
fig, ax = da1.plot2d(lw=1.5)
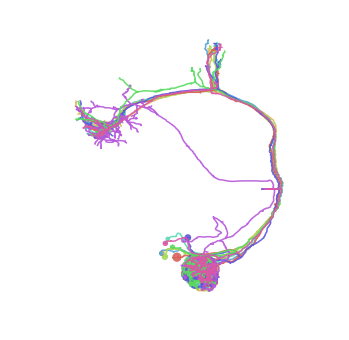
Fetching volumes¶
Lets also fetch a volume to put those neurons into context:
[18]:
vol = pymaid.get_volume('v14.neuropil')
vol
[18]:
<navis.Volume(name=v14.neuropil, color=(120, 120, 120, 0.6), vertices.shape=(8997, 3), faces.shape=(18000, 3))>
[25]:
# For visualization we rely on navis
import navis
# Make the volume more transparent
vol.color = (230, 230, 230, .2)
# Plot
fig, ax = navis.plot2d([da1, vol], lw=2)
# Rotate to frontal view
ax.elev = ax.azim = -90
ax.dist = 5
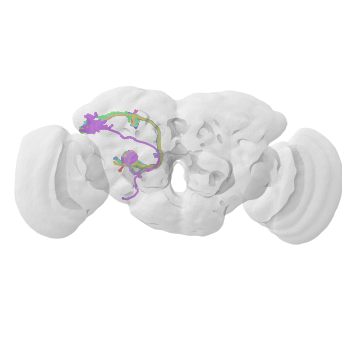
Obscure functions¶
Fetching skeletons and volumes is pretty standard, and fetching connectivity is covered in a separate tutorial. Let’s have a look at some more obscure functions.
Find out who contributed to the DA1 neurons we loaded above:
[31]:
cont = pymaid.get_contributor_statistics(da1)
# `cont` is a pandas.Series containing e.g. nodes contributed
# Let's plot only people with >1000 nodes contributed
{k: v for k, v in cont.node_contributors.items() if v > 1000}
[31]:
{'harrisonb': 2587,
'jefferis': 2031,
'schlegelp': 9977,
'koppenhaverb': 3355,
'ratliffj': 2177,
'hallouc': 3328,
'robertsr': 14537,
'masoodpanahn': 1436,
'kmecoval': 5203,
'alij': 3740,
'ranftp': 5930,
'fisherc': 2286,
'imtiazs': 2029,
'adamjohn': 2791,
'allredg': 1696,
'michaelLingelbach': 4253}
Have any of these nodes been imported (as opposed to being manually placed):
[33]:
iinfo = pymaid.get_import_info(da1)
iinfo
Fetching info: 100%|██████████| 1/1 [00:02<00:00, 2.37s/it]
[33]:
{'2863104': {'n_imported_treenodes': 1524},
'57381': {'n_imported_treenodes': 502},
'61221': {'n_imported_treenodes': 679},
'57353': {'n_imported_treenodes': 1005},
'57323': {'n_imported_treenodes': 951},
'755022': {'n_imported_treenodes': 1271},
'27295': {'n_imported_treenodes': 1161},
'1811442': {'n_imported_treenodes': 916},
'57311': {'n_imported_treenodes': 608}}
Looks like some parts of these neurons have been imported.
Last but not least, let’s check who has reviewed these neurons:
[38]:
rev = pymaid.get_review_details(da1)
# `rev` is a DataFrame that contains timestamps of when a user has reviewed a given node
rev.head(3)
[38]:
| node_id | imtiazs | masoodpanahn | kmecoval | fisherc | ratliffj | helmickl | alij | sharifin | robertsr | ... | meechank | edmondsona | jamasba | schlegelp | calles | hallouc | adesinaa | lovef | mooree | skeleton_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 3047214 | 2016-05-04 21:05:00 | NaT | NaT | NaT | NaT | NaT | NaT | NaT | NaT | ... | NaT | NaT | NaT | NaT | NaT | NaT | NaT | NaT | NaT | 2863104 |
| 1 | 3047212 | 2016-05-04 21:05:00 | NaT | NaT | NaT | NaT | NaT | NaT | NaT | NaT | ... | NaT | NaT | NaT | NaT | NaT | NaT | NaT | NaT | NaT | 2863104 |
| 2 | 3047210 | 2016-05-04 21:05:00 | NaT | NaT | NaT | NaT | NaT | NaT | NaT | NaT | ... | NaT | NaT | NaT | NaT | NaT | NaT | NaT | NaT | NaT | 2863104 |
3 rows × 24 columns
Scaling things up¶
So far, the examples have pulled only a small amount of data but pymaid scales really well. Let’s demonstrate how to fetch all neurons in the FAFB dataset, shall we?
First we need to compile a list of neurons above a certain size and with a soma using pymaid.find_neurons():
[3]:
all_fafb = pymaid.find_neurons(min_size=1000, only_soma=True)
INFO : Get all neurons with >= 1000 nodes (pymaid)
INFO : Found 7920 neurons matching the search parameters (pymaid)
all_fafb is now a list of 7920 neurons. At the moment these neurons only consist of skeleton ID and a name - any other data such as nodes or connectors are not (yet) loaded. Note the NAs:
[5]:
all_fafb[:5]
[5]:
| type | name | skeleton_id | n_nodes | n_connectors | n_branches | n_leafs | cable_length | soma | units | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CatmaidNeuron | PD5g1_L#3 8880132 ECM Tarly family left | 8880131 | NA | NA | NA | NA | NA | NA | 1 nanometer |
| 1 | CatmaidNeuron | KCaBx 12719 CF | 10190852 | NA | NA | NA | NA | NA | NA | 1 nanometer |
| 2 | CatmaidNeuron | aSP-g type tract crossing alpha lobe neuron 98... | 983054 | NA | NA | NA | NA | NA | NA | 1 nanometer |
| 3 | CatmaidNeuron | Uniglomerular mALT VA6 adPN 017 DB | 16 | NA | NA | NA | NA | NA | NA | 1 nanometer |
| 4 | CatmaidNeuron | Multiglomerular mALT l2PN VM1+VP2+VP3+23 LTS 0... | 32793 | NA | NA | NA | NA | NA | NA | 1 nanometer |
The not-yet-available data is lazily loaded on request:
[6]:
all_fafb[0].nodes.head()
[6]:
| node_id | parent_id | creator_id | x | y | z | radius | confidence | type | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 38680419 | 38680415 | 127 | 649397.0 | 128668.0 | 170760.0 | 51.0 | 5 | slab |
| 1 | 38680424 | 38680419 | 127 | 649497.0 | 128728.0 | 170840.0 | 51.0 | 5 | slab |
| 2 | 38680426 | 38680424 | 127 | 649733.0 | 128876.0 | 170920.0 | 51.0 | 5 | slab |
| 3 | 38680436 | 38680432 | 127 | 649945.0 | 129012.0 | 171040.0 | 51.0 | 5 | slab |
| 4 | 38680432 | 38680426 | 127 | 649901.0 | 128984.0 | 171000.0 | 51.0 | 5 | slab |
Note how the first neuron is now the only one with more detailed info in the summary:
[7]:
all_fafb[:5]
[7]:
| type | name | skeleton_id | n_nodes | n_connectors | n_branches | n_leafs | cable_length | soma | units | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | CatmaidNeuron | PD5g1_L#3 8880132 ECM Tarly family left | 8880131 | 3129 | 96 | 126 | None | 883607 | [38712262] | 1 nanometer |
| 1 | CatmaidNeuron | KCaBx 12719 CF | 10190852 | NA | NA | NA | NA | NA | NA | 1 nanometer |
| 2 | CatmaidNeuron | aSP-g type tract crossing alpha lobe neuron 98... | 983054 | NA | NA | NA | NA | NA | NA | 1 nanometer |
| 3 | CatmaidNeuron | Uniglomerular mALT VA6 adPN 017 DB | 16 | NA | NA | NA | NA | NA | NA | 1 nanometer |
| 4 | CatmaidNeuron | Multiglomerular mALT l2PN VM1+VP2+VP3+23 LTS 0... | 32793 | NA | NA | NA | NA | NA | NA | 1 nanometer |
So in theory, you now have about ~8k neurons to work with. But while the lazy loading is neat, it is also rather slow because it is done sequentially (i.e. one neuron at a time). If you want skeleton information (nodes + connectors) for all these neurons, you are best off triggering a full download which will use parallel threads:
[ ]:
all_fafb.get_skeletons()
By default, pymaid uses 100 parallel threads to fetch that data. On slow internet connections, this can cause errors. To reduce the maximum number of parallel threads, you can do this:
[ ]:
rm.max_threads = 10
That’s it for now. Check out the API reference for a full list of available functions.