pymaid.cluster_by_synapse_placement¶
- pymaid.cluster_by_synapse_placement(x, sigma=2000, omega=2000, mu_score=True, restrict_cn=None, remote_instance=None)[source]¶
Cluster neurons based on their synapse placement.
Distances score is calculated by calculating for each synapse of neuron A: (1) the Eucledian distance to the closest synapse in neuron B and (2) comparing the synapse density around synapse A and B. This is type-sensitive: presynapses will only be matched with presynapses, post with post, etc. The formula is described in Schlegel et al., eLife (2017):
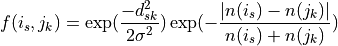
The synapse similarity score for neurons i and j being the average of
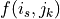 over all synapses s of i. Synapse k is the
closest synapse of the same sign (pre/post) in neuron j to synapse s.
over all synapses s of i. Synapse k is the
closest synapse of the same sign (pre/post) in neuron j to synapse s.
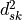 is the eucledian distance between these distances.
Variable
is the eucledian distance between these distances.
Variable 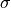 (
(sigma) determines what distance between s and k is considered “close”.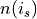 and
and 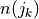 are
defined as the number of synapses of neuron i/j that are within given
radius
are
defined as the number of synapses of neuron i/j that are within given
radius  (
(omega) of synapse s and j, respectively (same sign only). This esnures that in cases of a strong disparity between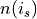 and
and 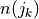 , the synapse similarity will be
close to zero even if the distance between s and k is very small.
, the synapse similarity will be
close to zero even if the distance between s and k is very small.- Parameters:
x –
Neurons as single or list of either:
skeleton IDs (int or str)
neuron name (str, exact match)
annotation: e.g. ‘annotation:PN right’
CatmaidNeuron or CatmaidNeuronList object
sigma (int, optional) – Distance in nanometer between synapses that is considered to be “close”.
omega (int, optional) – Radius in nanometer over which to calculate synapse density.
mu_score (bool, optional) – If True, score is calculated as mean between A->B and B->A comparison.
restrict_cn (int | list | None, optional) –
- Restrict to given connector types:
0: presynapses
1: postsynapses
2: gap junctions
3: abutting connectors
If None, will use all connectors. Use either single integer or list. E.g.
restrict_cn=[0, 1]to use only pre- and postsynapses.remote_instance (CatmaidInstance, optional) – If not passed, will try using globally defined. Need to provide if neurons are only skids or annotation(s).
- Returns:
Object that contains distance matrix and methods to plot dendrograms.
- Return type: